So you’ve just been accepted for publication. At this point, after months of extra experiments and back and forths with reviewers, you’re probably well and truly sick of your paper. However, the months roll by you look at the paper again, you note a cute bit of data analysis here, nice turn of phrase there, and with a gleam in your eye you look to see how many citations you’ve gotten. And unless you’re very lucky, that number might well be still in single digits. 12 months of work, and less than 10 people have ever cited your work. Maybe you feel like it was all for nothing. Well I’m here to make you feel better, because your work was much more important than that.
You see, maybe only a few people cited your work. But that was science that might very well have been impossible without your work. Those are a half dozen scientific children you’ve produced. And how many people then cited those papers, those papers that would have been impossible without you? That might very well be say another 6 for each of those, so that’s 36 scientific grand-children that would have been impossible without you! A total family tree of 42 papers. And it keeps going… those 36 new papers might have all been cited too. So because of your 12 months of work, and a mild 6 citations, you have been single handled responsible for the generation of 258 papers. Don’t trust my numbers? Well let’s check it out.
So in my last post, we interacted with PubMed, with nicely formed requests, and parsed back our nice, neat XML in a careful considered way. Well unfortunately, we need to stop that. I need to get who cited you, and that is something that a) isn’t available with the PubMed API and b) it isn’t available by PubMed at all. The best we can do is find papers that cite you in PubmedCentral. (Yes, Google Scholar does keep this information, but it has no API, and more problematically, it does not have unique paper identifiers, so trawling through papers gets really difficult). So the first trick is to make PubMed think that Python is a web browser. That is achieved with this, where we in essence inherit a class from urllib, alter it, and then jam it back in to the original class:
class AppURLopener(urllib.FancyURLopener):
version = 'Mozilla/5.0 (Windows; U; Windows NT 5.1; it; rv:1.8.1.11) Gecko/20071127 Firefox/2.0.0.11'
urllib._urlopener = AppURLopener()
The rest of the meat is in a recursive function. I really should have chopped this up into a few bits, so here is a slightly sanitized version so we can quickly understand what it does.
def whocitedme(pmid_in, cache):
#GET URL, GET DATA, PARSE TO TREE, THEN FIND ALL THE P TAGS
url = "http://www.ncbi.nlm.nih.gov/pubmed?linkname=pubmed_pubmed_citedin&from_uid=" + pmid_in
data = urllib.urlopen(url).read()
tree = ET.fromstring(data)
ps = tree.findall(".//{http://www.w3.org/1999/xhtml}p") #find all <p>s
pmids = [] #People who cited this paper
years = [] #Year of the paper
for elm in ps: #ITERATE OVER ALL THE P TAGS
if elm.attrib == {'class': 'title'}: #IF IT CONTAINS THE PMID
url_string = elm.find('{http://www.w3.org/1999/xhtml}a').get('href')
pmids.append(url_string[url_string.rfind('/')+1:]) #APPEND NEW PMIDS
if elm.attrib == {'class': 'details'}: #IF IT CONTAINS THE YEAR
for txt in elm.itertext():
if txt[2:6].isdigit(): #WHERE THE YEAR LIVES
years.append(txt[2:6]) #APPEND IT
new_ids = []
new_years = []
cache.append ( pmid_in )
if len(pmids) > 0: #IF WE FOUND SOMETHING
for pmid in pmids: #ITERATE OVER THE NEW PMIDS
if pmid not in cache: #IF WE HAVEN'T ALREADY FOUND THIS PAPER
super_new, super_year = whocitedme(pmid, cache) #RECURSE
if super_new is not None: #IF WE FOUND SOMETHING
new_ids.append( super_new )
new_years.append( super_year )
return new_ids + pmids, new_years + years
else: #BASE CASE
return None, None
And with that, and some cleaning up, we EVENTUALLY get back our data (we’re limited by the speed that PubMed responds, which is about once a second). When we think about our data, we can break out some calculus and make sense of it. Say at any one point in time the total size of our family tree (that is, the people who cited us, and the people who cited them, and so on) is F(t). How many citations each of those get should have little bearing on our original paper, but instead depend on the field we are in. If papers on average in our field get C citations each year, then the next year, we should have F(t) = F(t-1)*C + F(t-1) citations in our total family tree . The year after that, we should then have F(t) = (F(t-2)*C+F(t-2))*C + F(t-1) = F(t-1)*C + F(t-1) That is to say, that the increase in the size of our family tree every year is what we had last year times C, the field citability. Thus, we can make an update rule which says
Which can be solved to give us.
When t = 0, the exponent collapses to 1, and hence A must be equal to the number of citations in year 1. This puts us in an interesting position now. If we take a ratio of the number of citations we generate in a year, to the field citability C (that is, the average number of citations each paper in our field gets) then we get an unbiased measure of how good our paper is. That is to say, because certain fields generate very few citations (e.g. Mathematics) versus some that generate a lot (e.g. Biomedical Science) comparing a papers total citations is unfair. If this works out, then this might be an interesting way, not only to compare papers, but fields in the round.
But does it work out? Let’s look at some of my papers. The first two papers I ever worked on were basically identical fields: how the stargazer gene affects the expression of extrasynaptic GABAA receptors, but one was in the hippocampus and one was in the cerebellum. (REMEMBER: THIS IS ONLY FOR PAPERS IN PUBMEDCENTRAL!!!)
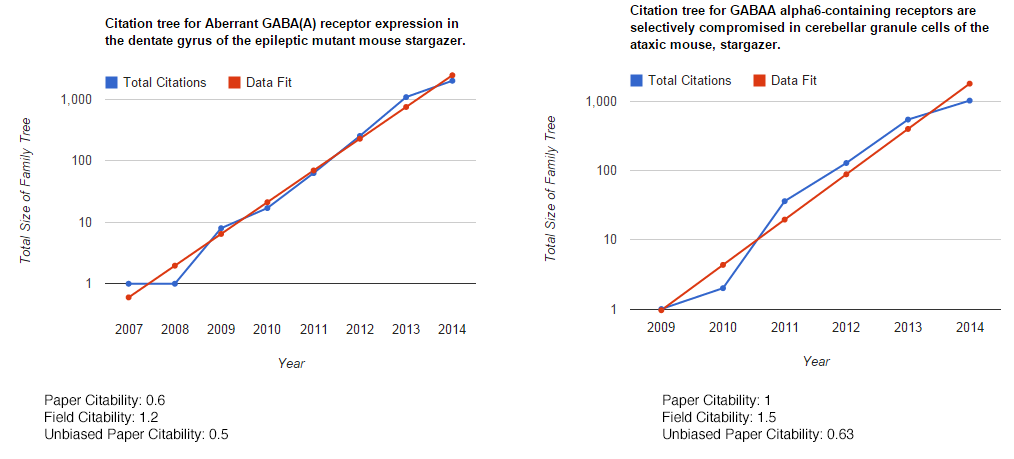
So we get very similar field citabilities for the two: 1.2 and 1.5. The unbiased citability turns out to be about the same, which is pretty good, as if we look at the total number of direct citations on Google Scholar, they’re about the same. As an aside, if we take the natural log of 2, and divide it by the field citability, it gives you the doubling time for you family tree, i.e. the family tree of those paper should double every 6 months.
How about two papers from two very different fields: In one paper I show for the first time that the histamine H4 receptor has an action on the brain. In the other, I wrote quite a nice story on the consequences of striatonigral and pallidonigral transmission. Both of these papers have had a bit of an impact, so what does my algorithm say?
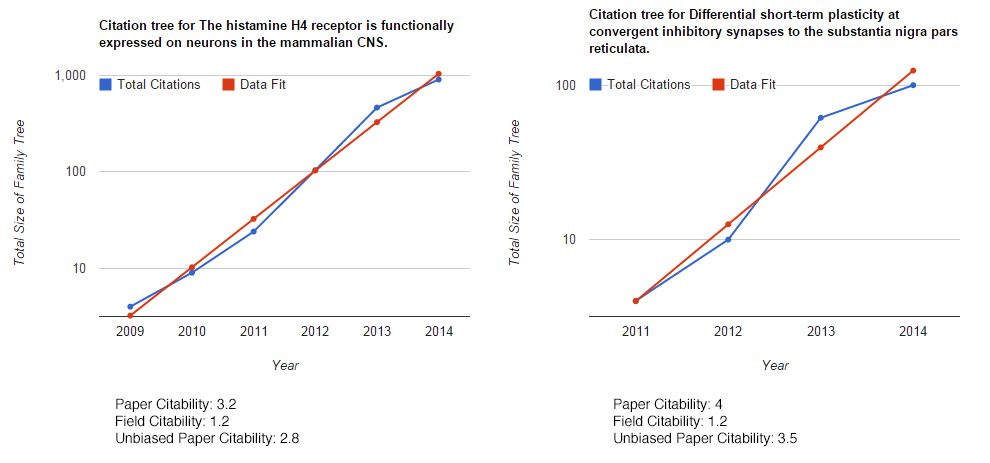
Well, we definitely see that the paper citability scales with my view of paper impact. However, I’m a bit surprised that the papers in the basal ganglia don’t generate more citations than those on the H4 receptor. However, the idea does work, if you don’t believe me, try putting 17353988 in the search box below, and you’ll see how citable the field of endoscopy is.
Anyway, why don’t you give it a try, and tell me how it works for you? Remember, it recursively searches through a lot of papers, so if you put in an old paper, it can take a really really long time (like half an hour). And if you’ve got no citations in PubMedCentral, it doesn’t return anything.
But ultimately, the point is this, even if your paper only gets a few citations, don’t underplay its impact. After only a few years, you might have helped thousands of papers be born into the world.
Full code available here
Thank you for your terrific posts! Are all your publications on PubMed? I wonder if someone has written a tool to get ACM or other databases’ citations.
All of my publications are on pubmed. What’s ACM? “Association for Computing Machinery”? With a name like that, I would hope they have a very friendly API.